| Type: | Package |
| Title: | Hazard of Multi-Stage Clonal Expansion Models |
| Version: | 1.0.2 |
| Date: | 2025-12-05 |
| Author: | Cristoforo Simonetto
|
| Maintainer: | Cristoforo Simonetto <cristoforo.simonetto@ph.tum.de> |
| Description: | Functions to calculate hazard and survival function of Multi-Stage Clonal Expansion Models used in cancer epidemiology. For the Two-Stage Clonal Expansion Model an exact solution is implemented assuming piecewise constant parameters, see Heidenreich, Luebeck, Moolgavkar (1997) <doi:10.1111/j.1539-6924.1997.tb00878.x>. Numerical solutions are provided for its extensions, see also Little, Vineis, Li (2008) <doi:10.1016/j.jtbi.2008.05.027>. |
| License: | GPL-2 | GPL-3 [expanded from: GPL (≥ 2)] |
| Imports: | Rcpp (≥ 1.0.3), RcppParallel (≥ 5.0.0) |
| LinkingTo: | Rcpp, RcppParallel |
| Encoding: | UTF-8 |
| RoxygenNote: | 7.3.3 |
| Suggests: | knitr, rmarkdown, testthat |
| VignetteBuilder: | knitr |
| NeedsCompilation: | yes |
| Packaged: | 2025-12-16 23:37:14 UTC; cristoforo |
| Repository: | CRAN |
| Date/Publication: | 2025-12-17 00:00:02 UTC |
msce: Hazard of Multi-Stage Clonal Expansion Models
Description
Functions to calculate hazard and survival function of Multi-Stage Clonal Expansion Models used in cancer epidemiology. For the Two-Stage Clonal Expansion Model an exact solution is implemented assuming piecewise constant parameters, see Heidenreich, Luebeck, Moolgavkar (1997) doi:10.1111/j.1539-6924.1997.tb00878.x. Numerical solutions are provided for its extensions, see also Little, Vineis, Li (2008) doi:10.1016/j.jtbi.2008.05.027.
Author(s)
Maintainer: Cristoforo Simonetto cristoforo.simonetto@ph.tum.de (ORCID)
Other contributors:
Jan Christian Kaiser [contributor]
Exemplary person year table
Description
Data set of fatal lung cancer in rats exposed to Radon at Pacific Northwest National Laboratory. For each rat, the age at start of Radon exposure, the age at end of exposure and the age at end of follow-up/death is provided in weeks. The Radon dose rate is given in WL (working level).
Usage
data(lungCancerRadon)
Format
Data frame with 5 variables.
Source
Pacific Northwest National Laboratory, see Heidenreich, W, Jacob P, Paretzke H., et al. (1999). Radiation research, 151 2, 209-17 DOI:10.2307/3579772.
Examples
data(lungCancerRadon)
lungCancerRadon[1000,]
Exemplary person year table
Description
Fictitious data set providing the number of lung cancer cases and person years in some population stratified according to age, age of smoking start, age of smoking cessation, and average number of cigarettes per day during the active smoking period.
Usage
data(lungCancerSmoking)
Format
Data frame with 6 variables.
Source
Fictitious data set, only for illustration and test.
Examples
data(lungCancerSmoking)
lungCancerSmoking[1000,]
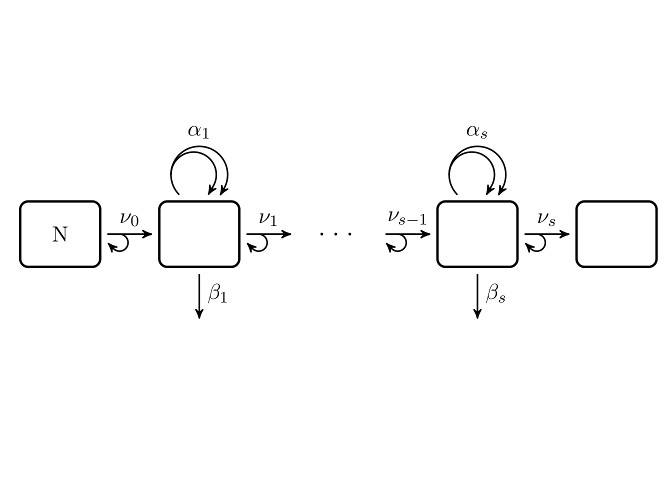
Numerical solution of the Multi-Stage Clonal Expansion Model
Description
This function aims to solve the general multi-stage model
with piecewise constant parameters approximatively
by integrating the characteristic equations with Euler's method.
For sufficiently small time intervalls, this approximation often
yields reasonable results.
Small time intervals can either be provided explicitly with many columns
in argument t.
An alternative is the optional parameter innerSteps.
Usage
msce_numerical(t, parameterList, innerSteps = 1000L)
Arguments
t |
Each element in a row of |
parameterList |
List of Matrices.
Each list member has to be named. Allowed names are Values in matrices |
innerSteps |
Positive integer.
To improve accuracy, each time interval is internally
divided into |
Details
Value
The output list contains all used arguments of the
parameterList and vectors of the model results for
hazard and logarithm of the survival function
for each row of the input.
See Also
Examples
t <-matrix(data=c(10,20,65,10,20,70),nrow=2,byrow=TRUE)
Nnu0 <- matrix(c(0.3,0.7,1),nrow = 1)
nu1 <- matrix(1e-6,nrow=1,ncol=3)
alpha1<- matrix(1,nrow=1,ncol=3)
gamma1<- matrix(c(0.13,0.13,0.13, 0.15,0.15,0.15),nrow=2,byrow=TRUE)
pars = list(Nnu0=Nnu0, nu1=nu1,alpha1=alpha1,gamma1=gamma1)
msce_numerical(t,pars)
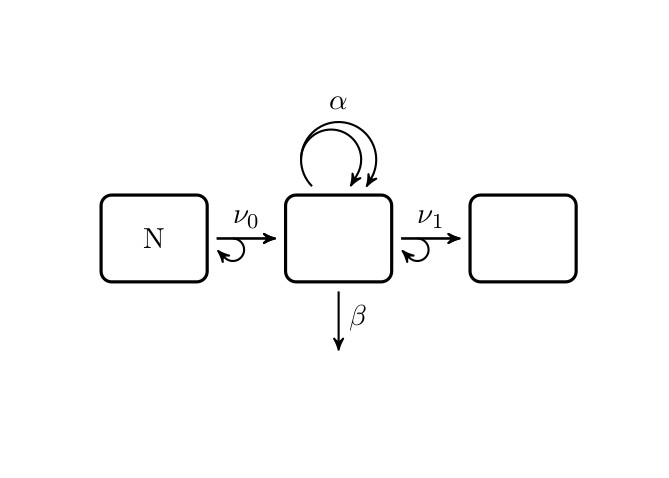
Exact solution of the Two-Stage Clonal Expansion Model
Description
For piecewise constant parameters tsce(t,parameterList)
returns the exact hazard and logarithm of the survival function
of the Two-Stage Clonal Expansion Model.
All arguments are matrices.
Evaluation is performed separately for each row.
Usage
tsce(t, parameterList)
Arguments
t |
Each element in a row of |
parameterList |
List of Matrices.
Each list member has to be named. Allowed names are Values in matrices |
Details
Value
Returns a list. Besides the parameters of the parameterList, it contains two named components: hazard the exact hazard and lnSurvival the logarithm of the survival function of the Two-Stage Clonal Expansion Model.
See Also
Examples
t <-matrix(data=c(10,20,65,10,20,70),nrow=2,byrow=TRUE)
Nnu0 <- matrix(c(0.3,0.7,1),nrow = 1)
alpha<- matrix(1,nrow=1,ncol=3)
gamma<- matrix(c(0.13,0.13,0.13, 0.15,0.15,0.15),nrow=2,byrow=TRUE)
nu1 <- matrix(1e-6,nrow=1,ncol=3)
pars = list(Nnu0=Nnu0, alpha=alpha,gamma=gamma,nu1=nu1)
tsce(t,pars)